Using Genetics to Predict Physical Traits
November 12, 2021 - 8 minutes readSeeing through DNA
Although the genome is vast and largely unexplored, we know a great deal about how genetic differences affect a dog’s physical appearance. In the human world, predictions about appearance are a major aspect of DNA forensics. Can we guess what your dog’s coat color will look like without seeing them? The short answer is yes to some degree!
The genetics of physical appearance was the first area of exploration dog geneticists embarked upon. What genes make a chocolate Labrador Retriever or a cocoa-colored French Bulldog? Spoiler alert: Not the same gene! What delineates a Papillon’s butterfly ears from the Phalene’s moth wings? And why is the Chihuahua so tiny while the Great Dane towers over other dogs?
A simple matter of inheritance
We can use the alleles (versions of genetic markers) that your dog carries to predict many “simple” physical traits. “Simple” here means controlled by a few genes, or even a single gene. These are also called Mendelian traits because they often follow the patterns of inheritance observed by geneticist-botanist-monk Gregor Mendel. Dominant traits require only one allele in order to be expressed; recessive traits require two copies in order for us to observe the trait in an individual dog.
The exact combination of coat colors and patterns in your dog is created by complicated interactions between different genes. Some traits can be masked by the genes for other coat traits. The genetic term that describes when the effects of one gene get masked by the effects of another is epistasis. For example, a Labrador Retriever might have the genetics to be chocolate and still be yellow. The only sign of the underlying epistasis will be their brown nose, and of course, their genes!
We call a dog’s alleles, inherited from their mother and father, their “genotypes.” We determine genotypes using an approach called low-pass sequencing. This allows us to assess millions of genetic variants across a dog’s genome at the scale we need to make new discoveries, but it comes with some caveats for individual trait analysis. Occasionally, a dog’s genotype may not be confidently called, and we remove these low confidence calls during quality control. Such quality control is a possible source of inaccuracy in trait predictions.
Machine learning for quantitative traits
Predictive models that use machine learning are all about taking a set of data, identifying patterns, and coming up with a solution. The features which models use to predict a trait are called predictors. In genetics, the predictors are your dog’s genotypes.
Working with Shirley Li and the great team at Darwin’s Ark, our team employed a “random forest” machine learning algorithm to predict each dog’s relative body size from ankle-high to hip-high. These methods can be applied to other quantitative traits as well (really, any trait that exists on a spectrum). The computer will build many 1,000s of decision trees, which take a random selection of genotypes, and output an average prediction for the trait from all these trees.
Existing data from dogs enrolled in Darwin’s Ark, whose owners were surveyed for their dog’s relative height, were used to define the height prediction model. We first defined “training” and “test” sets of data from dogs of known heights and genetic markers associated with height differences. The trees were built using the training dogs, and then tested on the inputs from test dogs. We then compared the true heights of the test dogs to their predicted heights. The model had excellent performance on mixed-breed dogs, purebred dogs, and dogs who were measured in-person by staff members.
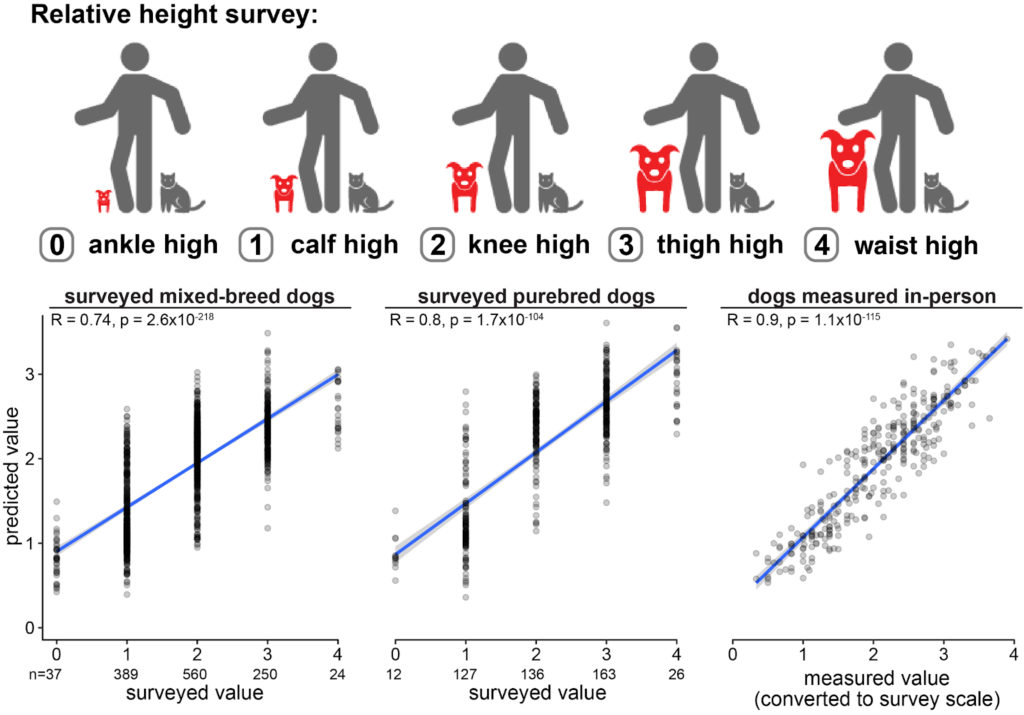 Prediction models have limitations. For example, this model does not account for the environmental aspects of growth, like prenatal and early life nutrition, and the prediction is for adult height, not considering a dog’s current age or growth trajectory. The model also performs best for heights more common in dogs: small to medium to large. Very tiny or very giant dogs are rare. In the Genomic Reports we provide for Foundation Cohort members, we provide several examples of breeds common among dogs at those surveyed heights.
Prediction models have limitations. For example, this model does not account for the environmental aspects of growth, like prenatal and early life nutrition, and the prediction is for adult height, not considering a dog’s current age or growth trajectory. The model also performs best for heights more common in dogs: small to medium to large. Very tiny or very giant dogs are rare. In the Genomic Reports we provide for Foundation Cohort members, we provide several examples of breeds common among dogs at those surveyed heights.
How far can predictions go?
Okay, so we can predict physical appearance with some ease. Does this mean that one day we’ll be able to predict a dog’s physiology, health, and behavior? Not for a while! Right now, genetic predictions are still very challenging to get right, even for people.
In the Foundation Cohort study, we provide information on two interesting, non-visual genetic traits; although, both are rather rare. Adaptation to high altitude hypoxia, an ability of Tibetan mastiffs, like their human companions, to tolerate low oxygen levels in the mountains, may be controlled by a stretch of DNA in the gene EPAS1, for which we test four markers. Shedding propensity, or the amount of fluff a dog is likely to set loose upon your furniture, is dependent on the gene MC5R and the “curly” coat gene, KRT71, and we test both markers to predict whether a dog has normal or low shedding. Your mileage may vary! These are predictions, after all, and no genetic prediction is 100% accurate.
The Future is Bright!
Our genetics team is incredibly excited to be working with the Foundation Cohort members at the Dog Aging Project. Together we expect to advance our knowledge of canine genetics by leaps and bounds!

Kathleen Morrill
Research Team