Making Sense of Genomic Sequence Data
July 14, 2021 - 10 minutes readFrom tall, lanky deerhounds to short, stocky terriers, dogs come in all shapes, sizes, and colors. One of the reasons that dogs are so fascinating is because they vary so widely from breed to breed. In fact, there are already over 550 breeds represented in the Dog Aging Project Pack as well as thousands of unique and wonderful mixed breed dogs. As part of the Foundation Cohort study, we are sequencing the genomes of 10,000 Pack members, and the first question we’ll be able to answer is the perpetual dog park query—What kind of a dog is that?
 Developing good reference panels
Developing good reference panels
The Foundation Cohort study will collect information about 40-60 million genetic variations in the canine genome for each dog. Most of these will be used to identify regions of the genome involved in disease, but about 680,000 of these markers can be used to make what we refer to as breed calls — the identification of genetic similarity to specific breed lineages, a proxy for ancestry in dogs from unrecorded family trees.
Seems like it should be easy to point to a little Cocker Spaniel DNA here or a little Malamute DNA over there, but in fact, making breed calls is pretty complex. Even defining what a breed is can be complicated. We could sequence a bunch of Corgis, for example, and identify distinct lineages among them. Which one of those lineages is the best representation of the breed? All of them? None?
This is where reference panels are extremely important! A reference panel refers to a collection of the genomic sequences of a sample of known-breed dogs. Thinking back to the Corgis, if we only included the genomic sequence of one Corgi, it wouldn’t be as useful as having a reference panel of five Corgis. If those sequences represented a broad range of Corgi lineages, they would be more useful representations of the breed as a whole, helping us to positively identify Corgis and Corgi mixes of unknown lineage.
Partnering Up!
For the Dog Aging Project, we collaborated with our partners at Darwin’s Ark, another community science project, to assemble reference panels that consist of twelve individual genomic sequences for each of 101 breeds. Most, but not all of these, are AKC-recognized breeds. We’re particularly grateful to the members of Darwin’s Ark who came together to help build the reference panels for American Pitbull Terriers and English Shepherds, two iconic breeds recognized by the United Kennel Club (UKC).
In developing these twelve-dog reference panels, we have tried to represent as many diverse lineages as possible. This gives us better accuracy when making matches between the reference panel and the sample DNA.
Sometimes participants ask us if they can share genetic information about their dog obtained from companies like Embark or Wisdom. The short answer is no. These DNA tests are fantastic for uncovering the mysteries of your dog’s ancestry and for testing markers linked to disease, but they apply genetic techniques that are different from the genome sequencing approach we use in Foundation Cohort study to make new discoveries.
 Making it all make sense
Making it all make sense
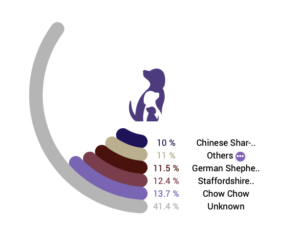
When we compare a dog’s genomic sequence to the 101 reference panels, we can infer two things: global ancestry and local ancestry. Global ancestry tells us what percentage of the dog’s DNA is similar to each of the breeds in the reference panel. Local ancestry reveals which specific portions of the dog’s DNA might have been inherited from different populations. We report global ancestry back to our participants with a circle plot like the one shown here.
For a purebred dog, we typically find that 80-100% of that dog’s genomic sequence matches with our reference panel for that breed. A lower percentage, like 80%, doesn’t mean that the dog isn’t really a purebred (breed calls are not a substitute for pedigree). It’s more likely that portions of the dog’s genetic material aren’t represented in our reference panel.
Similarly, our collection of breed-specific reference panels is not yet comprehensive. There are hundreds of dog breeds worldwide but we test for only 101 common breeds. If we analyze the genomic sequence of a purebred dog that is not represented, our analytical tools will often match it to a closely related breed. For example, a Flat-Coated Retriever might show up as a Chesapeake Bay Retriever or a Labrador Retriever.
In this example, 13.7% of the dog’s DNA is most similar to that of the reference panel for the Chow Chow with several other breeds represented at a similar level, but the majority of this dog’s genomic sequence couldn’t be matched to our reference panels. This can happen for several reasons. The ancestral breeds in this dog’s lineage might not be represented in our reference panels, or this mixed breed dog might not be descended from purebred dogs at all. The majority of dogs worldwide aren’t part of named breeds, and many beloved companion dogs represent this type of non-breed or “village” dogs.
Mix? Match? Mix-up?
Sometimes the results of our genomic analysis are surprising to owners, especially if our breed calls seem to contradict what owners have always known to be true about their dogs. Information about dogs comes from a wide variety of sources. Maybe we met our puppy’s parents. Maybe the shelter staff listed certain breeds on our adoption papers. Maybe we’ve made educated guesses about our dog’s ancestry based on physical characteristics.
It’s interesting how different kinds of information can lead us to different conclusions. Genomic testing isn’t always perfectly accurate for the reasons explained above, but often it does support drawing new and different conclusions about our dogs’ ancestry. Feedback from participants is important to us. After we share genomic results with participants, they have an opportunity to tell us what they think we got right or wrong in our analysis.
Foundation for what comes next
Making breed calls from genetic data is only one small part of the genetic research being conducted at the Dog Aging Project. Future posts in this series will focus on other aspects of our research, including analyzing genetic diversity, making predictions about physical traits from DNA, correlating health data with genetic data, and identifying the relative influence of environment and genetics on the aging process.
The Foundation Cohort study lays the foundation for many aspects of our research at the Dog Aging Project. We’re looking forward to many exciting discoveries in the years to come!

Kathleen Morrill
Research Team